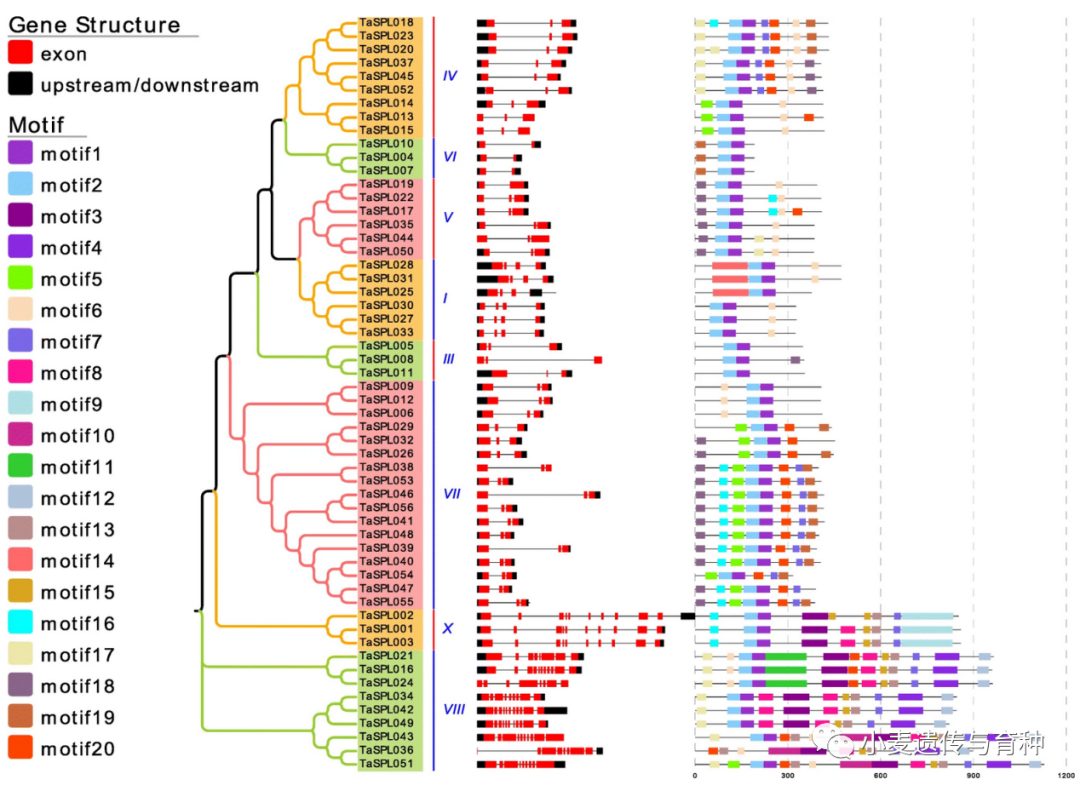
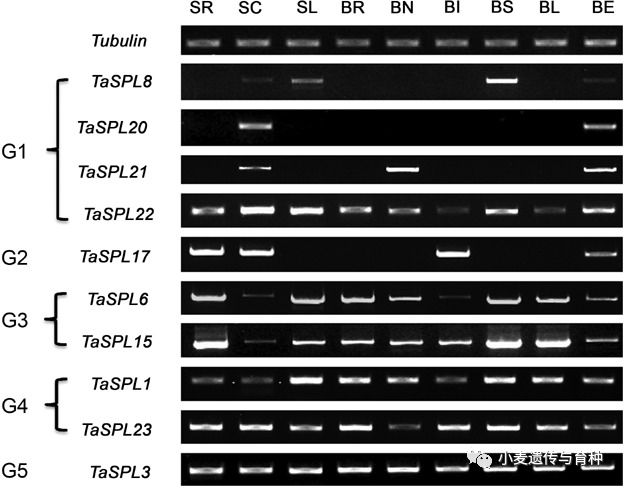
转录因子(TFs)是指能够以序列特异性方式结合DNA并且调节转录的蛋白质,参与植物生长发育的各个方面。SPL(SQUAMOSA promoter binding protein-like)是一类植物特有的转录因子,最初是在金鱼草中发现的,后来的研究发现该基因家族广泛存在于绿色植物中,SPL蛋白含有一个由78个氨基酸残基组成的高度保守的SBP 结构域,该结构域包含两个Zn2+结合位点以及一个核定位信号(NLS)。该结构域通过特异性结合基因启动子区域的顺式作用元件TNCGTACAA,从而调控SPL靶基因。目前在水稻中已发现多个OsSPL基因可影响穗,分蘖数,根系,籽粒形态等性状,例如水稻OsSPL3基因敲除株系表现为花期延迟,株高降低,穗长和粒长变短[1];敲除OsSPL6基因后,可影响水稻花序发育[2];水稻OsSPL7过表达植株分蘖数减少[3];OsSPL8基因可影响水稻叶型[4],OsSPL12基因可影响水稻根系发育[5],综上,说明了OsSPL基因在调控水稻生长发育和理想株型构建过程中发挥着重要的作用[1-7]。在水稻中,几乎每个OsSPL基因的功能通过构建转基因植株得到验证,但由于小麦基因组的复杂性,对SPL基因家族的研究较少。目前,通过比较基因组学方法发现小麦中共含有56个可能的TaSPL基因[8,9],但相关基因的鉴定、克隆和分子机制的研究较少,有待进一步研究。Zhang等人[10]从偃展4110中分离出10个TaSPL基因,分别是TaSPL1、TaSPL3、TaSPL6、TaSPL8、TaSPL15、TaSPL17、TaSPL20、TaSPL21、TaSPL22和TaSPL23,并通过系统进化分析将其分为G1到G5五类,G1中的TaSPL8/20/21和G2中的TaSPL17主要在茎尖分生组织和穗部表达水平较高;G3中的TaSPL6/15则在茎尖分生组织和穗部以外的其他组织表达水平较高,G4中的TaSPL1/23在不同组织间表达水平不一致;G5中的TaSPL3则在各组织间表达水平相同。Tissue‐specific expression patterns of TaSPL genes in nine tissues. The wheat tubulin gene was used as the control for equal cDNA concentrations. SR, SC, and SL, root, shoot apical meristem, and leaf blade at the seedling stage, respectively; BR, BN, BI, BS, BL, and BE, root, node, internode, leaf sheath, leaf blade, and ear at the late booting stage, respectively.Liu等人[11]利用快中子诱变获得了一个叶片直立、株型紧凑的小麦突变体,图位克隆结果表明该突变体是由于TaSPL8突变所致,进一步研究发现TaSPL8参与生长素和油菜素内酯生物合成途径来调控叶枕发育,使小麦叶夹角减小,株型更紧凑。
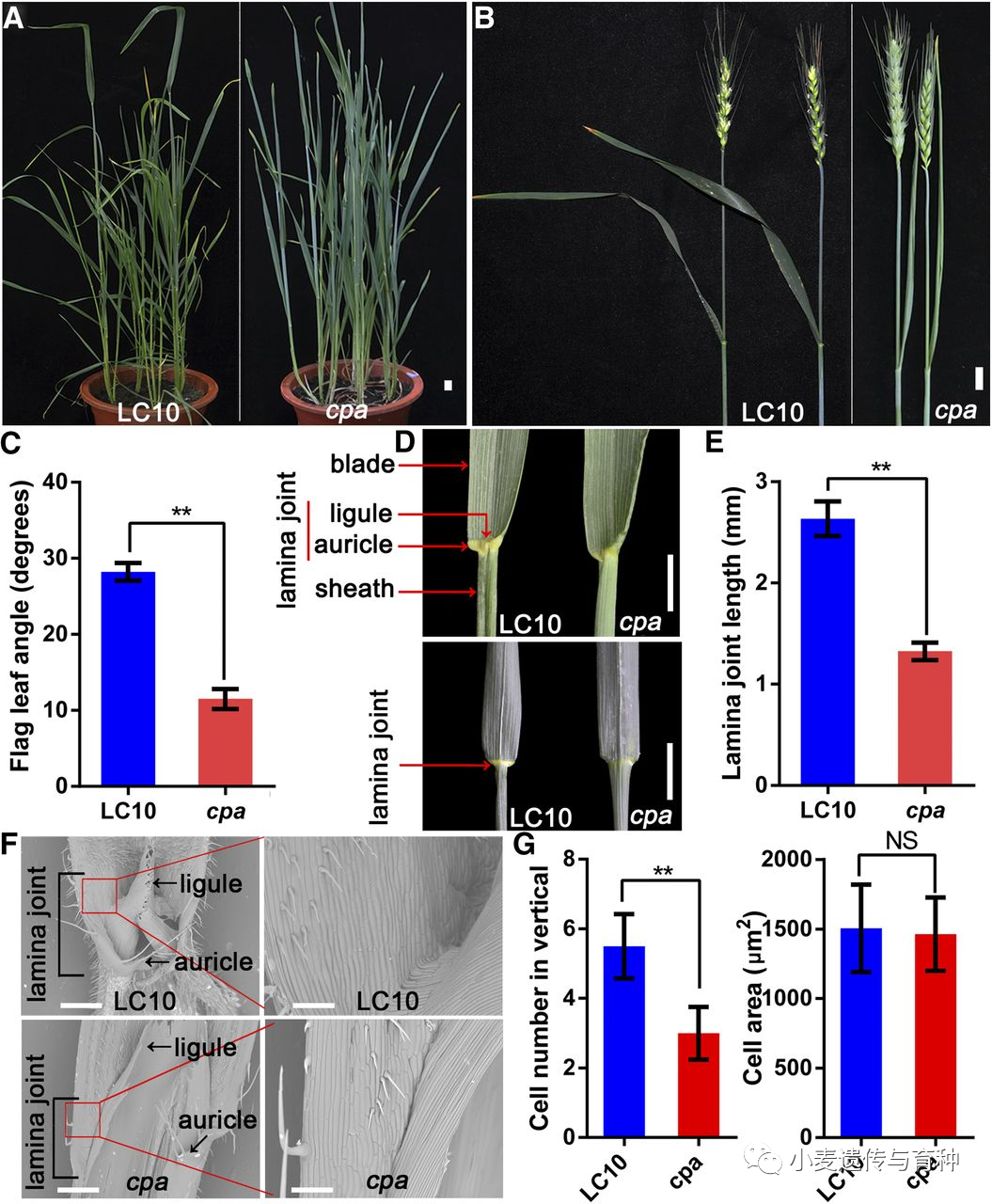 Morphological comparison of the wheat cultivar LC10 and the cpa mutant. A, Morphologies of LC10 and cpa plants at the heading stage. Bar = 2 cm. B, Morphologies of the flag leaf in LC10 and cpa plants at the flowering stage. Bar = 2 cm. C, Flag leaf angle of LC10 and cpa plants at the flowering stage. The double asterisk represents significant differences determined by the two-tailed Student's t test at P < 0.01. Error bars = sd (n = 10). D, The lamina joint phenotype of LC10 and cpa plants. Bars = 2 cm. E, Lamina joint length of LC10 and cpa plants. The double asterisk represents significant differences determined by two-tailed Student's t test at P < 0.01. Error bars = sd (n = 10). F, Morphology of the adaxial surface by a scanning electron microscope of the lamina joint of the LC10 and cpa flag leaf. Image magnification of the red box in the left (Bars = 200 μm) is shown on the right (Bars = 60 μm). G, Quantitative cell number and cell size of lamina joint from LC10 and cpa flag leaf. The cell number from the bottom to the top of the lamina joint was counted manually (n = 10) and cell size measured by the software ImageJ (n = 15). The double asterisk represents significant differences determined by two-tailed Student's t test at P < 0.01. Error bars = sd. NS, not significant.
Morphological comparison of the wheat cultivar LC10 and the cpa mutant. A, Morphologies of LC10 and cpa plants at the heading stage. Bar = 2 cm. B, Morphologies of the flag leaf in LC10 and cpa plants at the flowering stage. Bar = 2 cm. C, Flag leaf angle of LC10 and cpa plants at the flowering stage. The double asterisk represents significant differences determined by the two-tailed Student's t test at P < 0.01. Error bars = sd (n = 10). D, The lamina joint phenotype of LC10 and cpa plants. Bars = 2 cm. E, Lamina joint length of LC10 and cpa plants. The double asterisk represents significant differences determined by two-tailed Student's t test at P < 0.01. Error bars = sd (n = 10). F, Morphology of the adaxial surface by a scanning electron microscope of the lamina joint of the LC10 and cpa flag leaf. Image magnification of the red box in the left (Bars = 200 μm) is shown on the right (Bars = 60 μm). G, Quantitative cell number and cell size of lamina joint from LC10 and cpa flag leaf. The cell number from the bottom to the top of the lamina joint was counted manually (n = 10) and cell size measured by the software ImageJ (n = 15). The double asterisk represents significant differences determined by two-tailed Student's t test at P < 0.01. Error bars = sd. NS, not significant.Cao等人[12]克隆并鉴定了水稻OsSPL16在小麦中的同源基因TaSPL16,三个TaSPL16同源基因分别位于7A,7B,7D染色体上,都含有三个外显子和两个内含子。该基因在幼穗中表达水平较高,在营养组织中几乎检测不到该基因的表达,表明TaSPL16基因可能在小麦穗发育过程中起着重要作用。
分蘖数是小麦株型的重要组成部分之一,Zhang等人[13]研究发现TaSPL3/17能与TaPIL1相互作用,作者通过蛋白序列分析和互作实验发现TaPIL1能激活TaTB1的转录表达,并与同样能激活TaTB1转录的TaSPL3/17互作,从而调控小麦分蘖数。并且该结果还在水稻和拟南芥中得到验证,表明了转录因子PILs能和SPLs相互作用,在调控小麦分蘖和株型改良方面发挥重要作用。
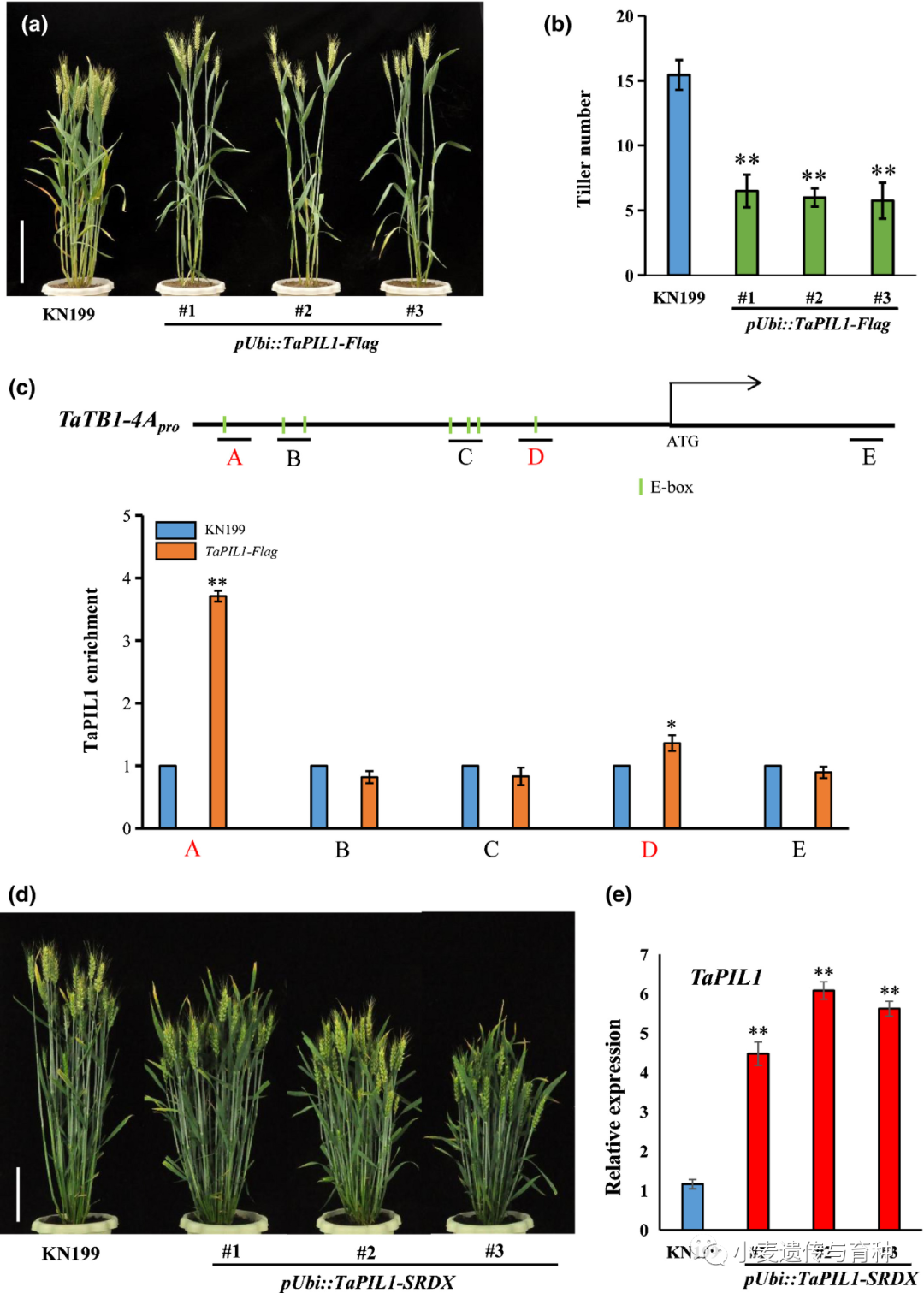 Wheat PHYTOCHROME‐INTERACTING FACTOR‐LIKE 1 (TaPIL1) associates with the promoter of TEOSINTE BRANCHED1‐4A (TaTB1‐4A). (a) Morphology of the TaPIL1‐Flag transgenic plants at the heading stage. Bar, 20 cm. (b) Tiller numbers and plant height of cv Kenong 199 (KN199) and TaPIL1‐Flag plants at the mature stage (n ≥ 15). Error bars denote ±SD (**, P < 0.01, Student’s t test). (c) Chromatin immunoprecipitation quantitative PCR (qPCR) showing the enrichment of TaPIL1 on the promoter of TaTB1‐4A. Schematic representation of different regions, which include the E‐box in the promoter of the TaTB1‐4A gene. Chromatin in TaPIL1‐Flag and KN199 stem base tissues at the jointing stage was immunoprecipitated using anti‐Flag antibodies. The precipitated DNA was analyzed by qPCR. Values represent the relative fold enrichment of the fragments between TaPIL1‐Flag and KN199. Error bars denote ±SD (*, P < 0.05; **, P < 0.01, Student’s t test). (d) TaPIL1 increases wheat tiller number by the addition of the SUPERMAN repression domain (SRDX) motif. Bar, 20 cm. (e) Expression levels of TaPIL1 in the KN199 and TaPIL1‐SRDX plants by quantitative reverse transcription PCR at the heading stage. The TaGAPDH gene was used as an internal reference. Error bars denote ±SD (**, P < 0.01, Student’s t test).
Wheat PHYTOCHROME‐INTERACTING FACTOR‐LIKE 1 (TaPIL1) associates with the promoter of TEOSINTE BRANCHED1‐4A (TaTB1‐4A). (a) Morphology of the TaPIL1‐Flag transgenic plants at the heading stage. Bar, 20 cm. (b) Tiller numbers and plant height of cv Kenong 199 (KN199) and TaPIL1‐Flag plants at the mature stage (n ≥ 15). Error bars denote ±SD (**, P < 0.01, Student’s t test). (c) Chromatin immunoprecipitation quantitative PCR (qPCR) showing the enrichment of TaPIL1 on the promoter of TaTB1‐4A. Schematic representation of different regions, which include the E‐box in the promoter of the TaTB1‐4A gene. Chromatin in TaPIL1‐Flag and KN199 stem base tissues at the jointing stage was immunoprecipitated using anti‐Flag antibodies. The precipitated DNA was analyzed by qPCR. Values represent the relative fold enrichment of the fragments between TaPIL1‐Flag and KN199. Error bars denote ±SD (*, P < 0.05; **, P < 0.01, Student’s t test). (d) TaPIL1 increases wheat tiller number by the addition of the SUPERMAN repression domain (SRDX) motif. Bar, 20 cm. (e) Expression levels of TaPIL1 in the KN199 and TaPIL1‐SRDX plants by quantitative reverse transcription PCR at the heading stage. The TaGAPDH gene was used as an internal reference. Error bars denote ±SD (**, P < 0.01, Student’s t test).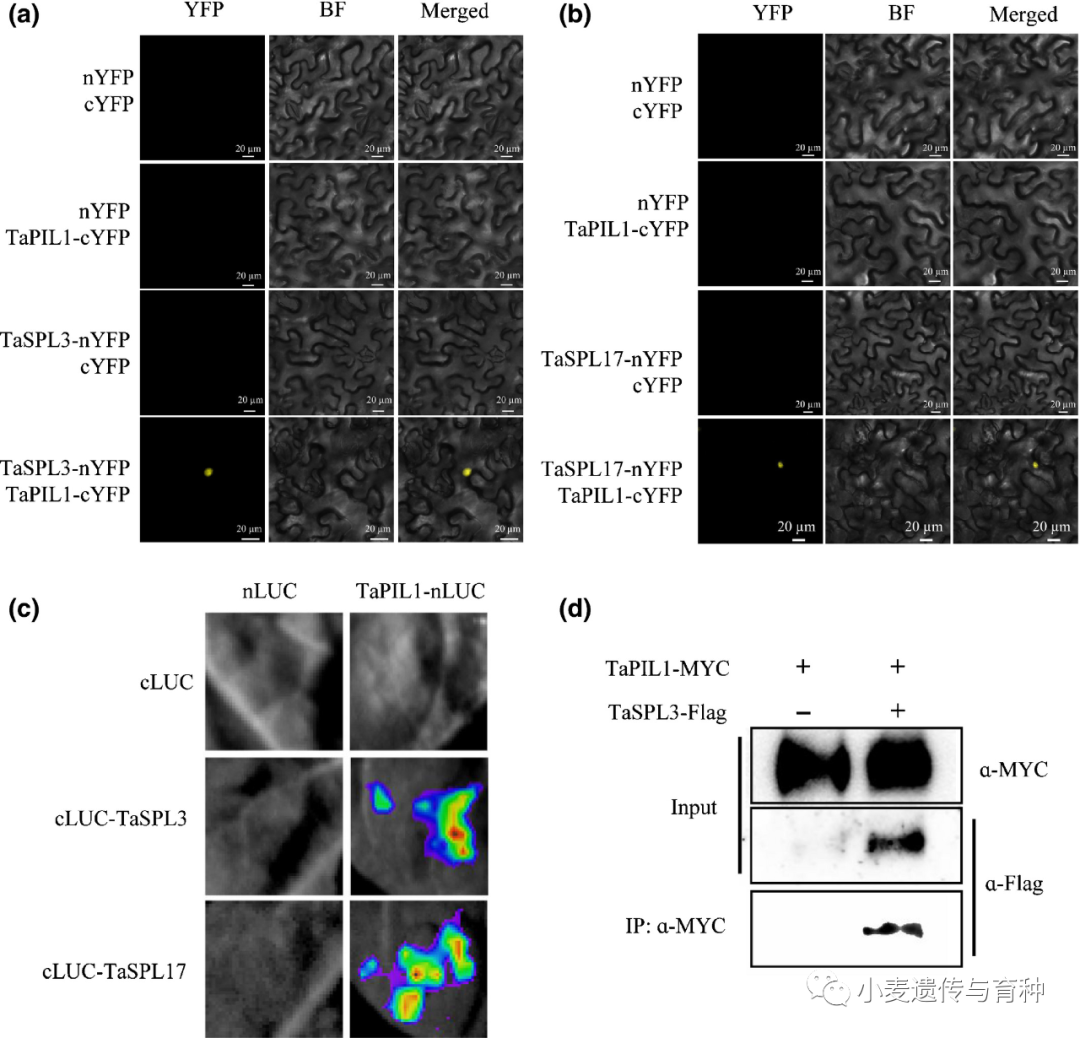 Physical interaction assays for wheat PHYTOCHROME‐INTERACTING FACTOR‐LIKE 1 (TaPIL1) and SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE (TaSPL)3/17. (a, b) Bimolecular fluorescence complementation assays of Nicotiana benthamiana leaves showing the interaction between TaPIL1 and TaSPL3/17. Bar, 20 μm. nYFP/cYFP, nYFP/TaPIL1‐cYFP, and TaSPL3‐nYFP/cYFP or TaSPL17‐nYFP/cYFP constructs were separately infiltrated into the upper left, upper right, and lower left parts of N. benthamiana leaves. The four coexpressed samples were used as the negative controls in this assay. The TaSPL3‐nYFP/TaPIL1‐cYFP and TaSPL17‐nYFP/TaPIL1‐cYFP constructs were infiltrated into the lower right quarter of corresponding N. benthamiana leaves, which were used as the experimental group. YFP, yellow fluorescent protein. (c) Luciferase (LUC) complementation imaging assays of N. benthamiana leaves showing the interaction between TaPIL1 and TaSPL3/17. (d) Coimmunoprecipitation assays of N. benthamiana leaves showing the interaction between TaPIL1 and TaSPL3. TaPIL1‐MYC and TaSPL3‐Flag were immunoprecipitated with anti‐MYC antibodies, and immunoblots were probed with anti‐MYC or anti‐Flag antibodies. IP, immunoprecipitation.
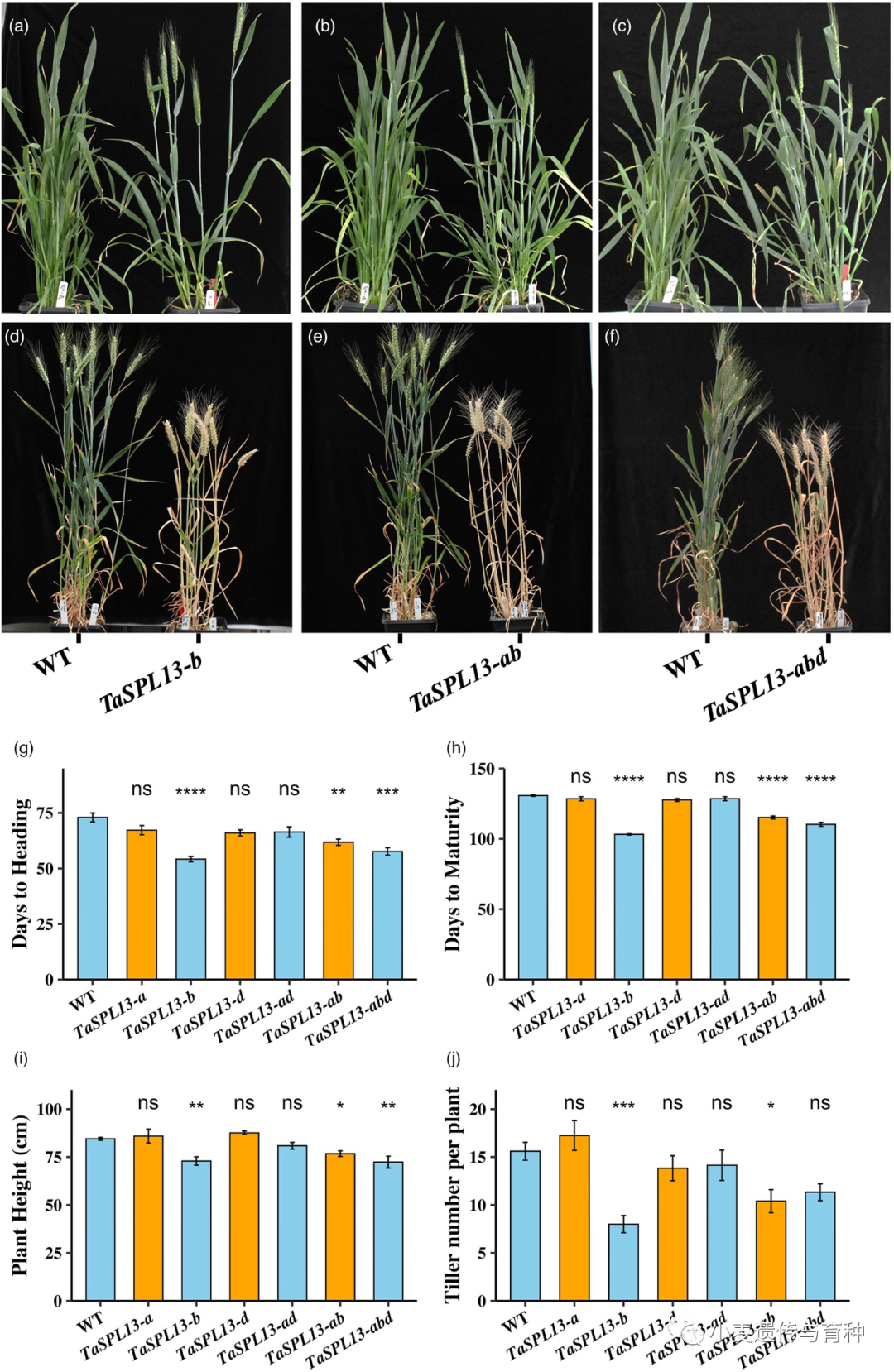
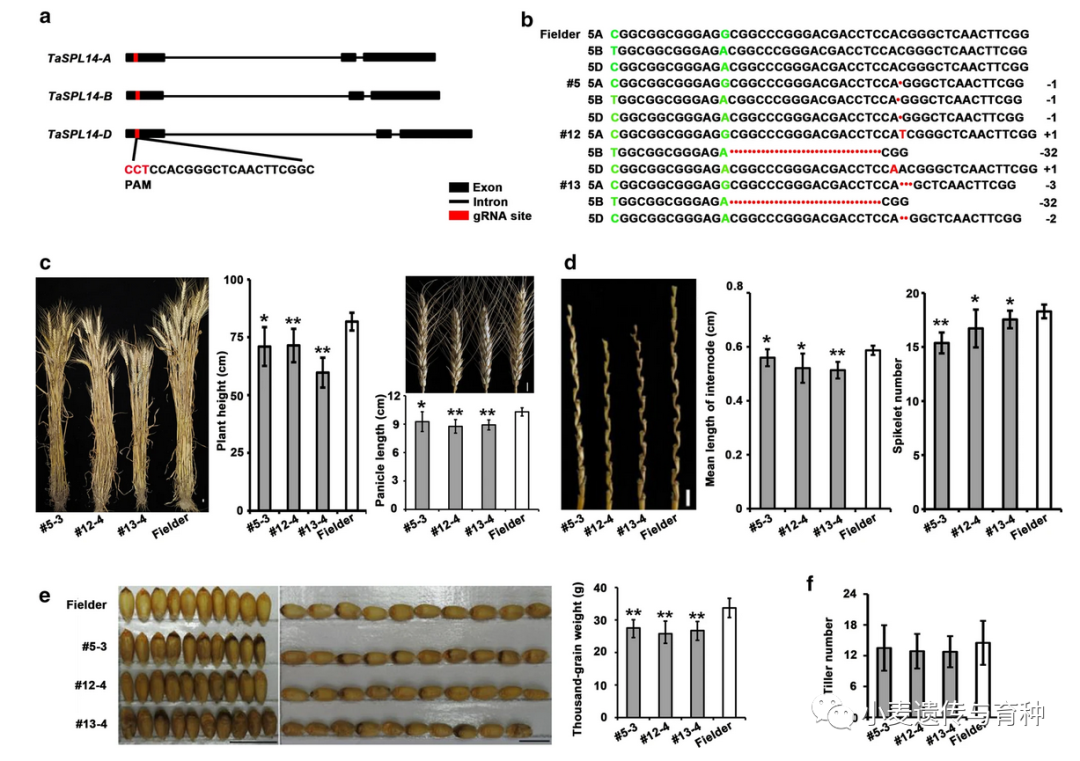
Physical interaction assays for wheat PHYTOCHROME‐INTERACTING FACTOR‐LIKE 1 (TaPIL1) and SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE (TaSPL)3/17. (a, b) Bimolecular fluorescence complementation assays of Nicotiana benthamiana leaves showing the interaction between TaPIL1 and TaSPL3/17. Bar, 20 μm. nYFP/cYFP, nYFP/TaPIL1‐cYFP, and TaSPL3‐nYFP/cYFP or TaSPL17‐nYFP/cYFP constructs were separately infiltrated into the upper left, upper right, and lower left parts of N. benthamiana leaves. The four coexpressed samples were used as the negative controls in this assay. The TaSPL3‐nYFP/TaPIL1‐cYFP and TaSPL17‐nYFP/TaPIL1‐cYFP constructs were infiltrated into the lower right quarter of corresponding N. benthamiana leaves, which were used as the experimental group. YFP, yellow fluorescent protein. (c) Luciferase (LUC) complementation imaging assays of N. benthamiana leaves showing the interaction between TaPIL1 and TaSPL3/17. (d) Coimmunoprecipitation assays of N. benthamiana leaves showing the interaction between TaPIL1 and TaSPL3. TaPIL1‐MYC and TaSPL3‐Flag were immunoprecipitated with anti‐MYC antibodies, and immunoblots were probed with anti‐MYC or anti‐Flag antibodies. IP, immunoprecipitation.Li等人[14]通过对水稻OsSPL基因的系统进化分析和比较,将56个小麦TaSPL基因分成八类;对部分TaSPL基因的表达模式进行验证发现,TaSPL1基因在叶片中表达水平较高,TaSPL7基因在穗部表达水平较高,TaSPL13基因在节间和籽粒中表达水平较高,TaSPL15/16基因在茎中表达水平较高。在所有组织的表达分析中,TaSPL13基因在穗中表达量最高,其小麦转基因植株的小花数和穗粒数显著增加。此外,Gupta等人[15]发现microRNA156识别元件(MRE)突变会导致TaSPL13转录本增加两倍左右,其突变体的开花期缩短、分蘖数减少和株高降低,籽粒大小及数量增加。这些结果表明TaSPL13基因有利于小麦理想株型构建,在育种改良方面具有很大潜力。
Effect of TaSPL13 mutations on flowering time and plant architecture during the 2020 fall season. (a–f) Phenotype of TaSPL13 mutant lines for heading and maturation time. The TaSPL13‐b/ab/abd plants (g) headed and (h) matured earlier compared with the WT plants, (i) plant height of TaSPL13‐b/ab/abd mutants reduced and (j) the tiller number also reduced. Error bars represent the standard error to the mean, * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, and **** indicates P < 0.0001. The P‐values are adjusted for multiple comparisons using Bonferroni correction水稻OsSPL14(也被称为IPA1)被称为一个新的“绿色革命”基因,该基因在调控水稻株型方面具有重要作用。研究发现,OsSPL14点突变影响了miR156对OsSPL14的定向调控,从而导致其分蘖数减少,抗倒伏性能和产量增加,此外还有研究发现该基因还能增强水稻抗病性,这些结果都表明OsSPL14对于水稻理想株型构建具有重要意义。Cao等人[16]对水稻OsSPL14基因在小麦中的同源基因TaSPL14进行了研究,结果表明小麦的TaSPL14基因敲除植株株高、穗长、小穗数和千粒重降低,与水稻中OsSPL14敲除植株的表型相似;但对分蘖数并无影响。转录组分析结果表明,相较于野生型,TaSPL14突变体幼穗中与乙烯反应相关基因的表达水平显著降低。前人研究证明TaSPL14能够直接结合到乙烯应答基因TaEIL1、TaRAP2.11和TaERF1的启动子上,并促进它们的表达,这表明TaSPL14可能通过乙烯响应途径来调控小麦穗发育。
Phenotypic analysis of TaSPL14 knock-out lines.SPL基因家族在调控植物生长发育方面具有重要作用,目前在水稻中已发现多个OsSPL基因在理想株型构建中起重要作用,其中OsSPL14基因更是被称为一个新的“绿色革命”基因。根据前人研究结果,小麦中存在56个TaSPL基因,受限于基因组的复杂性和突变体数量,目前仅对部分TaSPL基因进行鉴定克隆和分子机制的分析:TaSPL3/17能与TaPIL1相互作用调控小麦分蘖数;TaSPL8能影响叶枕发育,进而调控小麦叶夹角;TaSPL13在穗部表达水平较高,其MRE突变会影响株高、分蘖和籽粒大小等性状,这些基因对创制小麦理想株型有着重要影响,继续深入挖掘小麦TaSPL基因的调控机制,同时对已验证克隆的TaSPL基因资源,综合利用分子标记、基因编辑和传统育种手段,通过合理的基因聚合方案来进行小麦株型改良,以期培育出新的小麦高产品种,为我国粮食安全提供保障。
目的用于学术分享,转载注明来处,如有描述欠妥之处,敬请批评指正。若有侵权,请联系删除或修改。[1] Jiang, M.; He, Y.; Chen, X.; Zhang, X.; Guo, Y.; Yang, S.; Huang, J.; Traw, M.B. CRISPR-based assessment of genomic structure in the conserved SQUAMOSA promoter-binding-like gene clusters in rice. Plant J. 2020, 104, 1301–1314.[2] Wang, Q.L.; Sun, A.Z.; Chen, S.T.; Chen, L.S.; Guo, F.Q. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat. Plants 2018, 4, 280–288.[3] Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f–OsSPL7–OsGH3.8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130.[4] Lee, J.; Park, J.J.; Kim, S.L.; Yim, J.; An, G. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 2007, 65, 487–499[5] Shao, Y.; Zhou, H.Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 2019, 31, 1257–1275.[6] Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028.[7] Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544.[8] Zhu, T., Liu, Y., Ma, L. et al. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol 20, 420 (2020).[9] Guo, F.Y.; Lu, Q.W.; Cang, J. Genome-wide identification and expression profiling of the SPL family genes in wheat. Botany 2021,99, 185–198.[10] Zhang, B, Liu, X, Zhao, G, Mao, X, Li, A, Jing, R (2014) Molecular characterization and expression analysis of Triticum aestivum squamosa-promoter binding protein-box genes involved in ear development. J Integr Plant Biol 56: 571– 581.[11] Liu, K.; Cao, J.; Yu, K.; Liu, X.; Gao, Y.; Chen, Q.; Zhang, W.; Peng, H.; Du, J.; Xin, M.; et al. Wheat TaSPL8 modulates leaf angle through auxin and Brassinosteroid Signaling. Plant Physiol. 2019, 181, 179–194.[12] Cao, R.; Guo, L.; Ma, M.; Zhang, W.; Liu, X.; Zhao, H. Identification and functional characterization of squamosa promoter binding protein-like gene TaSPL16 in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 212.[13] Zhang, L., He, G., Li, Y., Yang, Z., Liu, T., Xie, X., Kong, X. and Sun, J. (2022), PIL transcription factors directly interact with SPLs and repress tillering/branching in plants. New Phytol, 233: 1414-1425.[14] Li, L.; Shi, F.; Wang, Y.; Yu, X.; Zhi, J.; Guan, Y.; Zhao, H.; Chang, J.; Chen, M.; Yang, G.; et al. TaSPL13 regulates inflorescence architecture and development in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 296, 110516.[15] Gupta, A., Hua, L., Zhang, Z., Yang, B. and Li, W. (2023), CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol. J, 21: 536-548.[16] Cao, J., Liu, K., Song, W. et al. Pleiotropic function of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE gene TaSPL14 in wheat plant architecture. Planta 253, 44 (2021).